HW 3 - More Dplyr
This homework is due Tuesday, Sep 26 at 11:59pm.
Getting Started
Go to the sta199-f23-1 organization on GitHub. Click on the repo with the prefix
hw3. It contains the starter documents you need to complete the homework assignment.-
Clone the repo and start a new project in RStudio. Reminder instructions for doing so are given here:
Click on the green CODE button, select Use SSH (this might already be selected by default, and if it is, you’ll see the text Clone with SSH). Click on the clipboard icon to copy the repo URL.
In RStudio, go to File ➛ New Project ➛Version Control ➛ Git.
Copy and paste the URL of your assignment repo into the dialog box Repository URL. Again, please make sure to have SSH highlighted under Clone when you copy the address.
Click Create Project, and the files from your GitHub repo will be displayed in the Files pane in RStudio.
Click hw-3.qmd to open the template Quarto file. This is where you will write up your code and narrative for the lab.
Packages
Exercise 1
Exercise 1 is a review of common questions. Below, list out the values (options) each code chunk argument can take on and define how the code chunk argument impacts our rendered document.
What does each of the following chunk options do? For the first four types listed below, also indicate which values they can take.
— eval
— error
— warning
— echo
— fig-height
— fig-width
Suppose we have the following tiny data frame:
# A tibble: 5 × 3
x y z
<int> <chr> <chr>
1 1 a K
2 2 b K
3 3 a L
4 4 a L
5 5 b K What does the code below do?
df |>
group_by(y)# A tibble: 5 × 3
# Groups: y [2]
x y z
<int> <chr> <chr>
1 1 a K
2 2 b K
3 3 a L
4 4 a L
5 5 b K What do the following pipelines do? Run both and analyze their results and articulate in words what each pipeline does. How are the outputs of the two pipelines different?
`summarise()` has grouped output by 'y'. You can override using the `.groups`
argument.# A tibble: 3 × 3
# Groups: y [2]
y z mean_x
<chr> <chr> <dbl>
1 a K 1
2 a L 3.5
3 b K 3.5# A tibble: 5 × 4
# Groups: y, z [3]
x y z mean_x
<int> <chr> <chr> <dbl>
1 1 a K 1
2 2 b K 3.5
3 3 a L 3.5
4 4 a L 3.5
5 5 b K 3.5Data: Mammals Sleep
The msleep (mammals sleep) data set contains the sleep times and weights for a set of mammals. This is a pre-loaded data set in R. A key for these data can be found below:
| column name | Description |
|---|---|
| name | common name |
| genus | taxonomic rank |
| vore | carnivore, omnivore or herbivore? |
| order | taxonomic rank |
| conservation | the conservation status of the mammal |
| sleep_total | total amount of sleep, in hours |
| sleep_rem | rem sleep, in hours |
| sleep_cycle | length of sleep cycle, in hours |
| awake | amount of time spent awake, in hours |
| brainwt | brain weight in kilograms |
| bodywt | body weight in kilograms |
Exercise 2
Which Didelphimorphia rank mammals sleep AT LEAST 16 hours? Answer the question using a single data wrangling pipeline. Your pipeline should present a data frame with just the variables name, order, and sleep_total, in descending order by their sleep total. Include a sentence listing the names of these mammals.
Exercise 3
What is the mean body weight and sleep total for any mammal that has monkey in their name? Answer the question using a single data wrangling pipeline. Your pipeline should present a data frame with two variables names mean_bw and mean_sp. (Hint: To act on rows of a data set based on a character string, please see hw-1 question 1 as an example.)
Suppose another researcher is interested in REM sleep for these mammals. However, this variable has missingness throughout. We want to know which order of mammal has the most missing values. Below, create a data frame with a single row that has a column with the name of the order, and a second column with the frequency of NAs values. Answer the question using a single data wrangling pipeline.
Exercise 4
Now, suppose a second research went out and collected the following information on more animals. Their data are titled msleep2.
When looking at their data, you notice that they have an entry for “Brown Fox”. You are not sure if Brown Fox is in the original msleep data. Write code to confirm or deny that Brown Fox is not an entry in msleep (Hint: does “Brown Fox” appear in the msleep dataset?). Write one sentence with your formal conclusion.
You are now asked to join msleep with msleep2. Below, write the code that will keep all of the rows in msleep that match with msleep2. Save this as a new data frame called new_data. Using in-line code, report the number of rows and columns in your new new_data data frame.
Now, we want to join msleep and msleep2 so that we include all rows from msleep and add columns from msleep2. Save this as a new data frame called new_data_b. Using in-line code, report the number of rows and columns in your new new_data_b data frame.
Exercise 5
Recreate the following plot using the msleep2 data set:
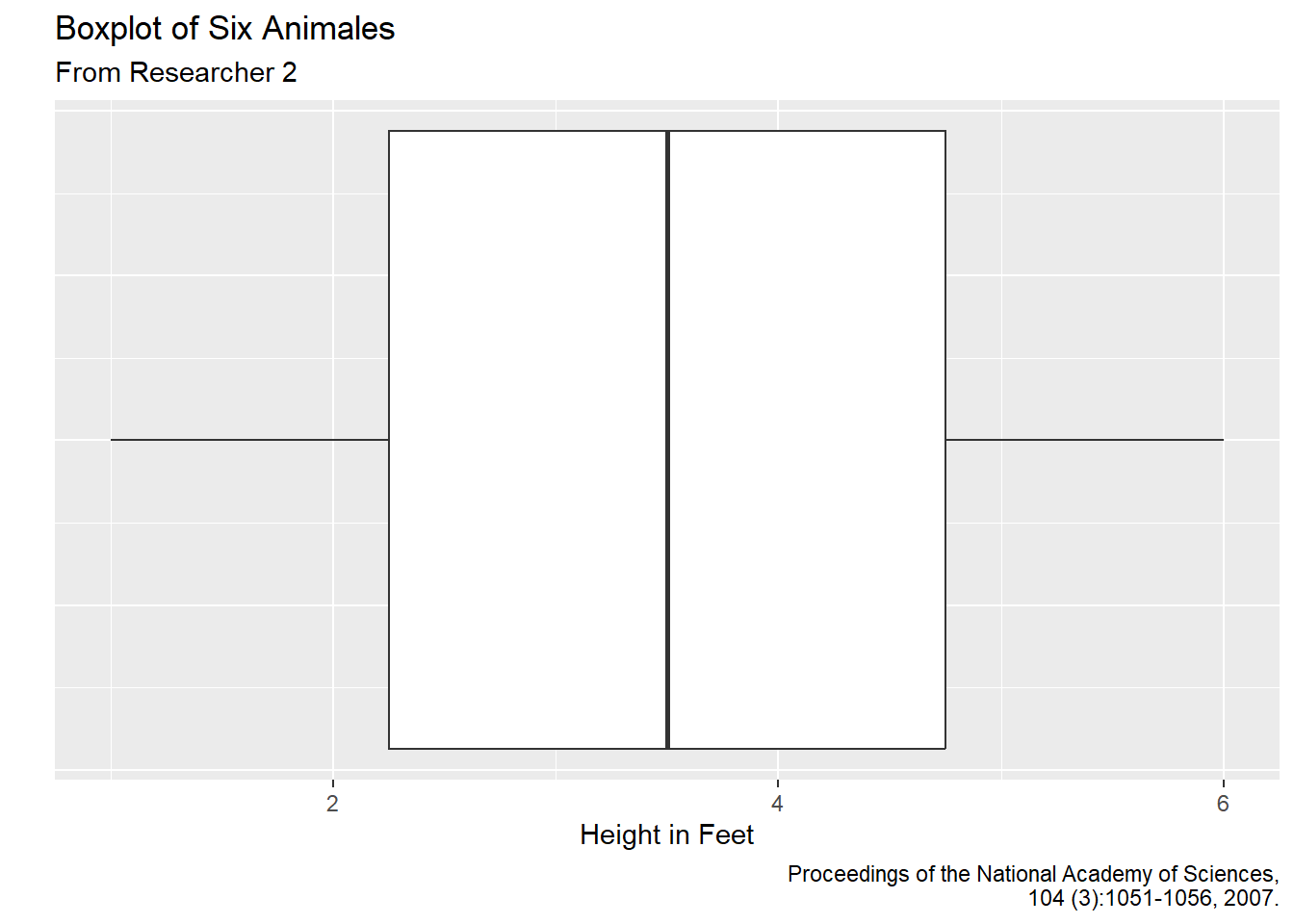
Hint: We can use functions within theme to removes text and tick marks from our axis. To reference the axis text, we can next axis.text.y within theme. To turn text off, we can set that = element_blank(): draws nothing, and assigns no space. Within theme use a similar method to also turn off the axis ticks. Note: You can reference the theme package here if needed.
What happens if you run as.numeric on name? In a new pipeline, try turning the variable name into a quantitative variable. Display your results and write 1-2 sentences on what happened.
Exercise 6
Part 1:
Please watch the following video on data visualization here .
After you have watched the entire video, please answer the following questions.
Fill in the blank aes with example code on how one might create the plot you see in the video (not including the animation). Note, you may choose your own variables names. The variables names you choose must be informative enough to be equated to what is seen in the video.
hans_rosling_data |>
ggplot(
aes(...)
) +
geom_point()Fun fact, if you want to incorporate something like this into your own scatter plots, you can learn about this here .
Part 2: Data Presentation
You will have a project presentation at the end of this semester and have to present data. Further, communicating data to a general audience is one of the most important skills one can develop. Reflecting on the video, please answer the following questions.
What is one way that Hans Rosling was effective in communicating the story of the data?
What is one strategy you may try and implement during your end of the year presentation? Why?
Submission
- Go to Gradescope and click Log in in the top right corner.
- Click School Credentials Duke Net ID and log in using your Net ID credentials.
- Click on your STA 199 course.
- Click on the assignment, and you’ll be prompted to submit it.
- Mark all the pages associated with exercise. All the pages of your homework should be associated with at least one question (i.e., should be “checked”). If you do not do this, you will be subject to lose points on the assignment.
- Do not select any pages of your PDF submission to be associated with the “Workflow & formatting” question.
Grading
Exercise 1: 5 points
Exercise 2: 5 points
Exercise 3: 9 points
Exercise 4: 8 points
Exercise 5: 8 points
Exercise 6: 10 points
Workflow + formatting: 5 points
Total: 50 points
